Core Data Phases for Intelligent Interoperability
Sign up for news and updates in population health
Jul 28, 2016
Population health management (PHM) and value-based care (VBC) are based on large amounts of patient data. The ability to aggregate health system data from all disparate sources, scrub it for quality and then analyze it for accuracy are the critical steps for the intelligent exchange of data – all the way to the end point of quality reporting to payers. For optimal success, these three core functions should be integrated within a software system.
Data Aggregation
Data aggregation should occur from all disparate EHRs being used within a given health network, done through a series of interfaces and data loads. Systems should be able to manage multiple formats, including HL7 transactions and C-CDA documents, in addition to supporting all major means of data transport, such as XDS.b, Direct and FHIR. Interoperability at a population level requires automation, flexibility and scale. The ability to aggregate is a critical first step, but only the beginning of assembling and reporting data at the necessary levels to impact care plans and satisfy advanced and alternative payment models built on risk.
Data Quality
The focus for data quality is two-fold. First, healthcare networks must understand their business goals in order to focus on the relevant data. Intelligent systems can use those goals to identify the relevant data and measure the data quality using key performance indicators and visualizations including interface integrity, provider attribution, patient duplicates, mapping accuracy and data completeness. Incoming data sets are monitored for consistency and accuracy, and alerts should be present to identify errors before they show up in reports and analytics. An essential part of data quality is to ensure completeness prior to submitting quality measures. For a quality measure score to be accurate, complete data from all sources must be aggregated and normalized into the measure components to generate a score.
Data Science
Data science is an emerging expertise whereby aggregated and scrubbed data is modeled to suggest gaps in care impacting both delivery and reimbursement. Here are three examples of data modeling Wellcentive currently performs.
CMS uses HCCs (Hierarchical Condition Categories) to tag patients with medical conditions, usually long-term conditions that impact the patient’s need for care. These codes are driven by the claims submitted to CMS. We prototyped a model to detect cases where a patient is missing an HCC code. For example, where a patient has HbA1C results, or insulin orders, but no diabetes HCC code, the model suggests that this code may be missing, a decision that would alter the patient’s care plan, but also reimbursement levels. Inversely, if the patient has an HCC for heart failure but no evidence in the data – no claims, no medications, no mention in notes – the model would suggest to review the case and see if there is a coding mistake or incomplete documentation.
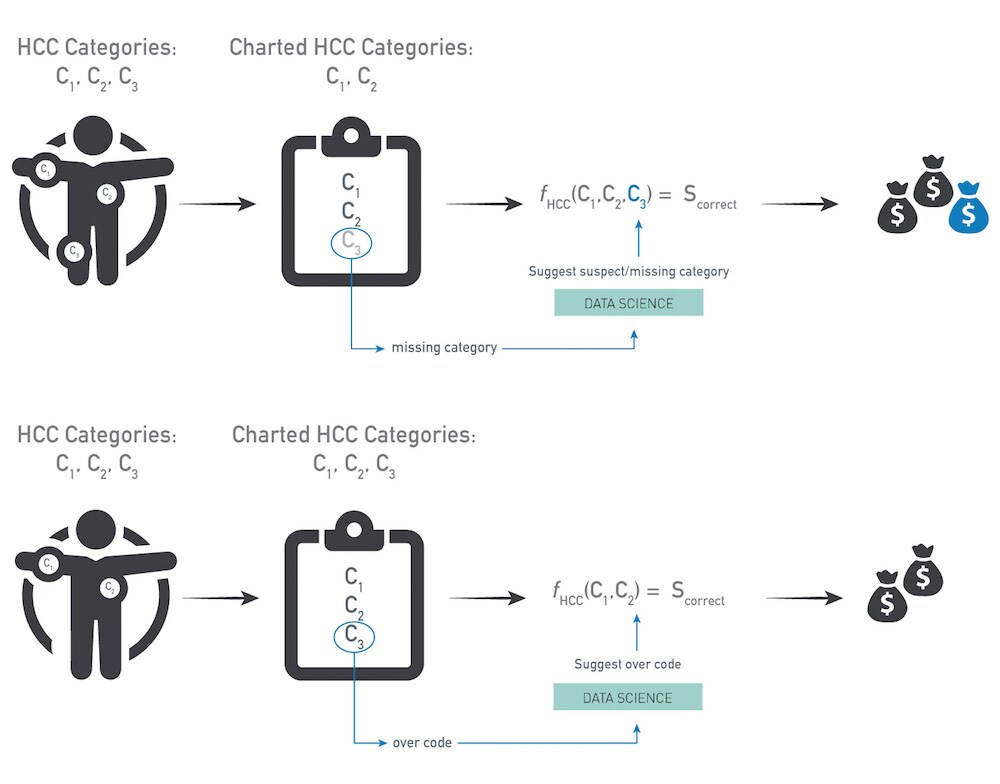
To build the HCC model, more than 35 million longitudinal patient charts were assembled over the last decade. The model can be made available to health networks as a set of HCC over/under coding tables for use in reports, care management work lists and practice profiles.
While standard terminologies have been available for many years, and are part of the meaningful use requirements, for example, the reality is that in most health network data sets from EHRs, approximately half the data uses standard codes, but the other half uses proprietary codes. This calls for developing a semi-automated mapping process: for all inbound data with non-standard codes (lab results, procedures, medications, problems, etc.) the system can propose a mapping from any source terminology to any target terminology including a LOINC code, CPT, SNOMED or RxNorm code. Once proprietary codes are mapped to known coding systems and/or value sets, the data will fit into the appropriate hierarchies and can be used in reports and queries. We developed the mapping algorithm by applying state-of-the-art machine learning and natural language processing techniques. The mappings were validated and benchmarked against the Unified Medical Language System (UMLS). Our mapping compendium now contains more than 4.5 million mappings – compared with the UMLS’ 800,000 mapped terms. This enables us to interpret the codes for large sets of proprietary data without a manual mapping effort.
External data sources are also useful to a health network’s database. For example, data sets with social determinants of health, geographical data and external benchmark data. Merging in external sources includes connecting a health network’s financial and clinical database, and weaving it in at the patient level. Importing external data sources allows networks to compare patient populations based on nearness to health facilities, socio-economic factors and other external parameters – the so-called “community vital signs.”[i] This is a lot to consider at the provider and health network level, but as value-based care evolves into population-based payment models, abilities around data aggregation, quality and science are becoming essential.
About the author

Kirk Elder,
PHM Global Innovation Leader, Philips
Kirk Elder is the PHM Global Innovation Leader at Philips. He is a 17-year veteran of the internet driven economy. He has held senior technology leadership positions at various companies at the forefront of revolutionary business models. He is passionate about fixing the healthcare financial crisis. Integrity, courage, and simplification form his core values.
You are about to visit a Philips global content page
Continue